
Spike Detection in Water Treatment Data in the R Language

Peter Prevos |
782 words | 4 minutes
Share this content
Spike detection in time series is essential to water treatment plant control. Spikes in SCADA data are events in the data stream of water treatment plants or similar installations. These spikes can indicate problems with the process and could result in an increased risk to public health. The WSAA Health Based Targets Manual specifies a series of decision rules to assess the performance of filtration processes. For example, this rule evaluates the performance of conventional filtration:
Individual filter turbidity ≤ 0.2 NTU for 95% of the month and not > 0.5 NTU for ≥ 15 consecutive minutes.
Turbidity is a measure for the cloudiness of fluid because of large numbers of individual particles otherwise invisible to the naked eye. Turbidity is an essential parameter in water treatment because a high level of cloudiness strongly correlates with the presence of microbes. This article uses the R language to show how to implement this specific decision rule.
Simulation
I first create a simulated SCADA feed for turbidity to create a minimum working example. The turbidity data frame contains 24 hours of data. The seq.POSIXt() function creates 24 hours of timestamps at a one-minute spacing. In addition, the rnorm() function creates 1440 turbidity readings with an average of 0.1 NTU and a standard deviation of 0.01 NTU. The image below visualises the simulated data. The next step is to assess this data following the decision rule.
# Simulate turbidity data
set.seed(1234)
turbidity <- data.frame(DateTime = seq.POSIXt(as.POSIXct("2017-01-01 00:00:00"),
by = "min", length.out = 24 * 60),
Turbidity = rnorm(n = 24*60, mean = 0.1, sd = 0.01)
)The second section simulates five spikes in the data. The first line picks a random start time for the spike. The second line in the for-loop picks a duration between 10 and 30 minutes. In addition, the third line simulates the value of the spike. The mean value of the spike is determined by the rbinom() function to create either a low or a high spike. The remainder of the spike simulation inserts the new data into the turbidity data frame.
## Spike detection
## Simulate data
for (i in 1:5) {
time <- sample(turbidity$DateTime, 1)
duration <- sample(10:30, 1)
value <- rnorm(1, 0.5 * rbinom(1, 1, 0.5) + 0.3, 0.05)
start <- which(turbidity$DateTime == time)
turbidity$Turbidity[start:(start+duration - 1)] <- rnorm(duration, value, value/10)
}
library(ggplot2)
ggplot(turbidity, aes(x = DateTime, y = Turbidity)) +
geom_line(size = 0.2) +
geom_hline(yintercept = 0.5, col = "red") +
ylim(0,max(turbidity$Turbidity)) +
theme_bw(base_size = 10) +
ggtitle("Simulated SCADA data")
ggsave("spikes.png", width = 4, height = 3)The image below visualises the simulated data using the mighty ggplot2 package. Only four spikes are visible because two of them overlap. The next step is to assess this data following the decision rule.
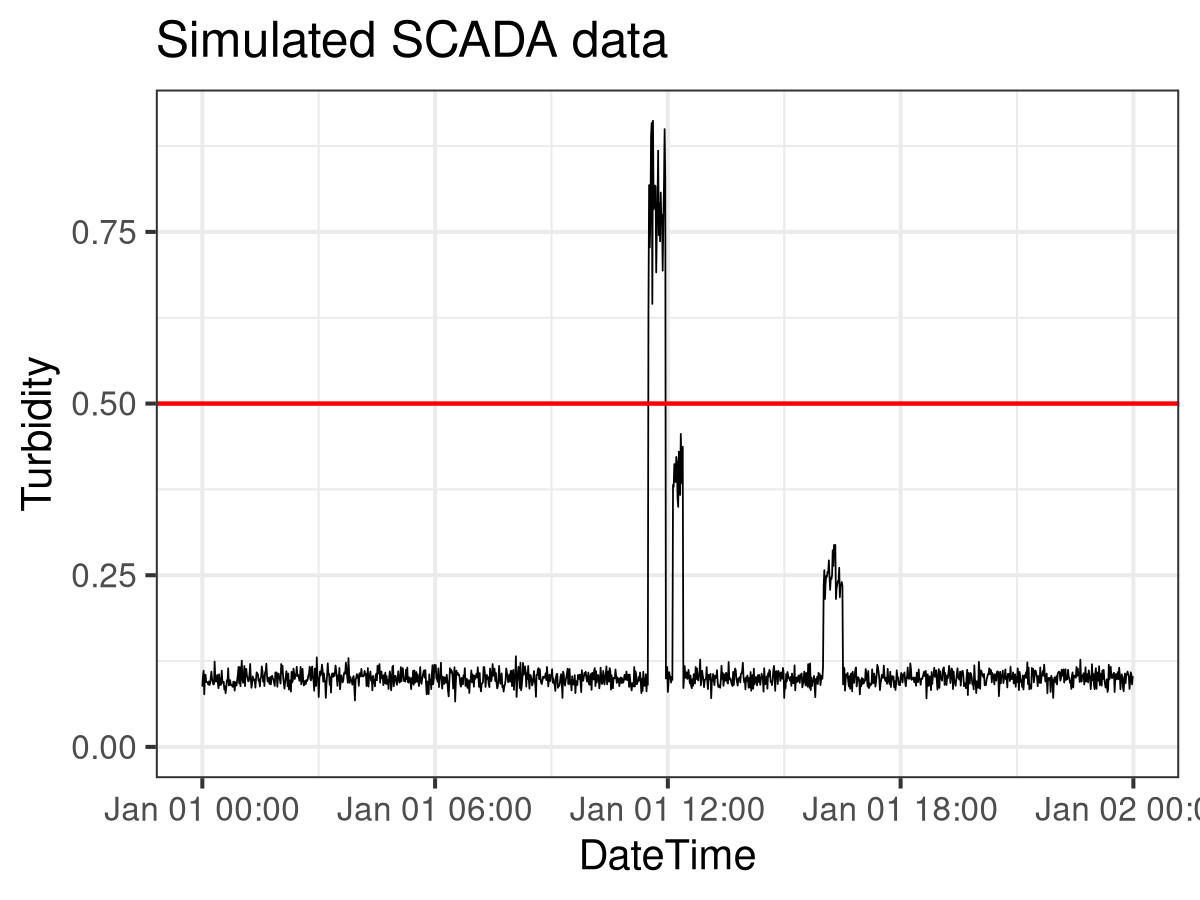
SCADA Spike Detection
The following code searches for all spikes over 0.50 NTU using the runlength function. This function transforms a vector into a vector of values and lengths. For example, the run length of the vector c(1, 1, 2, 2, 2, 3, 3, 3, 3, 5, 5, 6) is:
lengths: int [1:5] 2 3 4 2 1values : num [1:5] 1 2 3 5 6
The value 1 has a length of 1. The value 2 has a length of 3, and so on. The spike detection code creates the run length for turbidity levels greater than 0.5, which results in a boolean vector. The cumsum function calculates the starting point of each spike which allows us to calculate their duration.
The code results in a data frame with all spikes higher than 0.50 NTU and longer than 15 minutes. The spike at 11:29 was higher than 0.50 NTU and lasted 24 minutes. The other three spikes are either lower than 0.50 NTU. The first high spike lasted less than 15 minutes.
# Spike Detection Function
spike.detect <- function(DateTime, Value, Height, Duration) {
runlength <- rle(Value > Height)
spikes <- data.frame(Spike = runlength$values,
times = cumsum(runlength$lengths))
spikes$Times <- DateTime[spikes$times]
spikes$Event <- c(0,spikes$Times[-1] - spikes$Times[-nrow(spikes)])
spikes <- subset(spikes, Spike == TRUE & Event > Duration)
return(spikes)
}
spike.detect(turbidity$DateTime, turbidity$Turbidity, 0.5, 15)This approach was used for prototyping a software package to assess water treatment plant data following the Health-Based Targets Manual. The finished product has been written in SQL and is available under an Open Source sharing license.
The book Data Science for Water Utilities discusses anomaly detection in more detail.

Data Science for Water Utilities
Data Science for Water Utilities published by CRC Press is an applied, practical guide that shows water professionals how to use data science to solve urban water management problems using the R language for statistical computing.
Share this content


